Example Notebook: protein-ligand-benchmark
Related Publication
The preprint on “Best practices for constructing, preparing, and evaluating protein-ligand binding affinity benchmarks” provides accompanying information to this benchmark dataset and how to use it for alchemical free energy calculations. For any suggestions of improvements please raise an issue in its GitHub repository.
[1]:
from plbenchmark import targets
from IPython.core.display import HTML
Warning: Unable to load toolkit 'OpenEye Toolkit'. The Open Force Field Toolkit does not require the OpenEye Toolkits, and can use RDKit/AmberTools instead. However, if you have a valid license for the OpenEye Toolkits, consider installing them for faster performance and additional file format support: https://docs.eyesopen.com/toolkits/python/quickstart-python/linuxosx.html OpenEye offers free Toolkit licenses for academics: https://www.eyesopen.com/academic-licensing
Get the whole set of targets in the dataset
[2]:
# it is initialized from the `plbenchmark/sample_data/targets.yml` file
target_set = targets.TargetSet()
# to see which targets are available, one can get a list of names
target_set.get_names()
[2]:
['mcl1_sample']
The TargetSet is a Dict, but can be converted to a pandas.DataFrame or a html string via TargetSet.get_dataframe(columns=None) or TargetSet.get_html(columns=None). The default None for columns means that all columns are printed. One can also define a subset of columns as a list:
[3]:
HTML(target_set.get_html(columns=['name', 'fullname', 'pdb', 'references', 'numLigands', 'minDG', 'maxDG', 'associated_sets']))
/home/dhahn3/miniconda3/envs/plbenchmark/lib/python3.9/site-packages/pandas/core/dtypes/cast.py:1638: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
result[:] = values
[3]:
| name | fullname | pdb | references | numLigands | minDG | maxDG | associated_sets | |
|---|---|---|---|---|---|---|---|---|
| 0 | mcl1_sample | Induced myeloid leukemia cell differentiation protein Mcl-1 | 4HW3 | {'calculation': ['10.1021/ja512751q', '10.1021/acs.jcim.9b00105'], 'measurement': None} | 15 | -9.0 kilocalorie / mole | -6.1 kilocalorie / mole | [Schrodinger JACS] |
A target can be accessed with its name in two ways
[4]:
mcl1 = target_set['mcl1_sample']
mcl1_2 = target_set.get_target('mcl1_sample')
The Target class
contains all the available information about one target of plbenchmark. It also has two member variables, _ligand_set and _edge_set, which contain the information about the available ligand and edges of the respective target. A Target can either be accessed from the TargetSet (see cell before) or initialized using its name via
[5]:
mcl1 = targets.Target('mcl1_sample')
# The data in the column is stored in a pandas.Series and can be accessed via
mcl1.get_dataframe(columns=None)
/home/dhahn3/miniconda3/envs/plbenchmark/lib/python3.9/site-packages/pandas/core/dtypes/cast.py:1638: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
result[:] = values
[5]:
associated_sets [Schrodinger JACS]
comments hydrophobic interactions contributing to binding
date 2020-08-21
fullname Induced myeloid leukemia cell differentiation ...
id 99
ligands [lig_23, lig_26, lig_27, lig_28, lig_29, lig_3...
name mcl1_sample
netcharge xx
pdb 4HW3
references {'calculation': ['10.1021/ja512751q', '10.1021...
numLigands 15
maxDG -6.1 kilocalorie / mole
minDG -9.0 kilocalorie / mole
std(DG) 0.9 kilocalorie / mole
calculation REP1http://dx.doi.org/10.1021/ja512751qREP2Wan...
pdblinks REP1http://www.rcsb.org/structure/4HW3REP24HW3...
dtype: object
Access to the EdgeSet and LigandSet in different formats is achieved by
[6]:
mcl1_ligands = mcl1.get_ligand_set()
mcl1_ligands_df = mcl1.get_ligand_set_dataframe()
HTML(mcl1.get_ligand_set_html(columns = ['name', 'ROMol', 'measurement', 'DerivedMeasurement']))
[6]:
| name | ROMol | measurement | DerivedMeasurement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| comment | error | type | unit | value | Reference | type | value | error | unit | |||
| 0 | lig_23 | Table 2, entry 23 | 30 nanomolar | ki | nanomolar | 370 nanomolar | Friberg et al., J. Med. Chem. 2013 | dg | -8.83 kilocalorie / mole | 0.05 kilocalorie / mole | None | |
| 1 | lig_26 | Table 2, entry 26 | 0.44 micromolar | ki | micromolar | 1.0 micromolar | Friberg et al., J. Med. Chem. 2013 | dg | -8.24 kilocalorie / mole | 0.26 kilocalorie / mole | None | |
| 2 | lig_27 | Table 3, entry 27 | 0.0071 millimolar | ki | millimolar | 0.035 millimolar | Friberg et al., J. Med. Chem. 2013 | dg | -6.12 kilocalorie / mole | 0.12 kilocalorie / mole | None | |
| 3 | lig_28 | Table 3, entry 28, manually converted | 0.03 kilocalorie / mole | dg | kilocalorie / mole | -6.62 kilocalorie / mole | Friberg et al., J. Med. Chem. 2013 | dg | -6.62 kilocalorie / mole | 0.03 kilocalorie / mole | None | |
| 4 | lig_29 | Table 3, entry 29, manually converted | 120.0 calorie / mole | dg | calories / mole | -6940.0 calorie / mole | Friberg et al., J. Med. Chem. 2013 | dg | -6.94 kilocalorie / mole | 0.12 kilocalorie / mole | None | |
| 5 | lig_30 | Table 3, entry 30, manually converted | 0.6 micromolar | ic50 | micromolar | 1.9 micromolar | Friberg et al., J. Med. Chem. 2013 | dg | -7.85 kilocalorie / mole | 0.19 kilocalorie / mole | None | |
| 6 | lig_31 | Table 3, entry 31, manually converted | 80 nanomolar | ic50 | nanomolar | 1700 nanomolar | Friberg et al., J. Med. Chem. 2013 | dg | -7.92 kilocalorie / mole | 0.03 kilocalorie / mole | None | |
| 7 | lig_32 | Table 3, entry 32, manually converted | 0.08 dimensionless | pic50 | dimensionless | 4.8 dimensionless | Friberg et al., J. Med. Chem. 2013 | dg | -6.59 kilocalorie / mole | 0.11 kilocalorie / mole | None | |
| 8 | lig_33 | Table 3, entry 33, manually converted | 0.75 kilojoule / mole | dg | kilojoules / mole | -28.79 kilojoule / mole | Friberg et al., J. Med. Chem. 2013 | dg | -6.880975143403441 kilocalorie / mole | 0.17925430210325047 kilocalorie / mole | None | |
| 9 | lig_34 | Table 3, entry 34 | 3.2 micromolar | ki | micromolar | 9.9 micromolar | Friberg et al., J. Med. Chem. 2013 | dg | -6.87 kilocalorie / mole | 0.19 kilocalorie / mole | None | |
| 10 | lig_35 | Table 3, entry 35 | 0.14 micromolar | ki | micromolar | 0.38 micromolar | Friberg et al., J. Med. Chem. 2013 | dg | -8.81 kilocalorie / mole | 0.22 kilocalorie / mole | None | |
| 11 | lig_36 | Table 3, entry 36 | 0.1 micromolar | ki | micromolar | 1.1 micromolar | Friberg et al., J. Med. Chem. 2013 | dg | -8.18 kilocalorie / mole | 0.05 kilocalorie / mole | None | |
| 12 | lig_37 | Table 3, entry 37 | 0.15 micromolar | ki | micromolar | 0.3 micromolar | Friberg et al., J. Med. Chem. 2013 | dg | -8.95 kilocalorie / mole | 0.3 kilocalorie / mole | None | |
| 13 | lig_38 | Table 3, entry 38 | 2.1 micromolar | ki | micromolar | 7.7 micromolar | Friberg et al., J. Med. Chem. 2013 | dg | -7.02 kilocalorie / mole | 0.16 kilocalorie / mole | None | |
| 14 | lig_39 | Table 3, entry 39 | 0.7 micromolar | ki | micromolar | 7.6 micromolar | Friberg et al., J. Med. Chem. 2013 | dg | -7.03 kilocalorie / mole | 0.05 kilocalorie / mole | None | |
[7]:
mcl1_edges = mcl1.get_edge_set()
mcl1_edges_df = mcl1.get_edge_set_dataframe()
HTML(mcl1.get_edge_set_html())
/home/dhahn3/miniconda3/envs/plbenchmark/lib/python3.9/site-packages/pandas/core/dtypes/cast.py:1638: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
result[:] = values
[7]:
| ligand_a | ligand_b | name | Mol1 | Smiles1 | Mol2 | Smiles2 | exp. DeltaG [kcal/mol] | exp. Error [kcal/mol] | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | lig_28 | lig_35 | edge_28_35 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3C([H])([H])[H])[H])[H])[H])[H])[H])[H] | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)[H])[H])[H])[H] | -2.19 kilocalorie / mole | 0.22 kilocalorie / mole | ||
| 1 | lig_30 | lig_27 | edge_30_27 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])[H])[H])[H])[H])[H] | [H]c1c(c(c(c(c1[H])[H])OC([H])([H])C([H])([H])C([H])([H])C2=C(N(c3c2c(c(c(c3[H])[H])[H])[H])[H])C(=O)[O-])[H])[H] | 1.73 kilocalorie / mole | 0.22 kilocalorie / mole | ||
| 2 | lig_31 | lig_35 | edge_31_35 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C(F)(F)F)[H])[H])[H])[H])[H] | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)[H])[H])[H])[H] | -0.89 kilocalorie / mole | 0.22 kilocalorie / mole | ||
| 3 | lig_33 | lig_27 | edge_33_27 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])[H])Cl)[H])[H])[H])[H] | [H]c1c(c(c(c(c1[H])[H])OC([H])([H])C([H])([H])C([H])([H])C2=C(N(c3c2c(c(c(c3[H])[H])[H])[H])[H])C(=O)[O-])[H])[H] | 0.76 kilocalorie / mole | 0.22 kilocalorie / mole | ||
| 4 | lig_35 | lig_33 | edge_35_33 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)[H])[H])[H])[H] | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])[H])Cl)[H])[H])[H])[H] | 1.93 kilocalorie / mole | 0.28 kilocalorie / mole | ||
| 5 | lig_35 | lig_37 | edge_35_37 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)[H])[H])[H])[H] | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)C([H])([H])[H])[H])[H])[H] | -0.14 kilocalorie / mole | 0.37 kilocalorie / mole | ||
| 6 | lig_39 | lig_32 | edge_39_32 | [H]c1c(c(c(c(c1[H])[H])c2c(c(c(c(c2[H])[H])OC([H])([H])C([H])([H])C([H])([H])C3=C(N(c4c3c(c(c(c4[H])[H])[H])[H])[H])C(=O)[O-])[H])[H])[H])[H] | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])[H])C([H])([H])[H])[H])[H])[H])[H] | 0.44 kilocalorie / mole | 0.12 kilocalorie / mole |
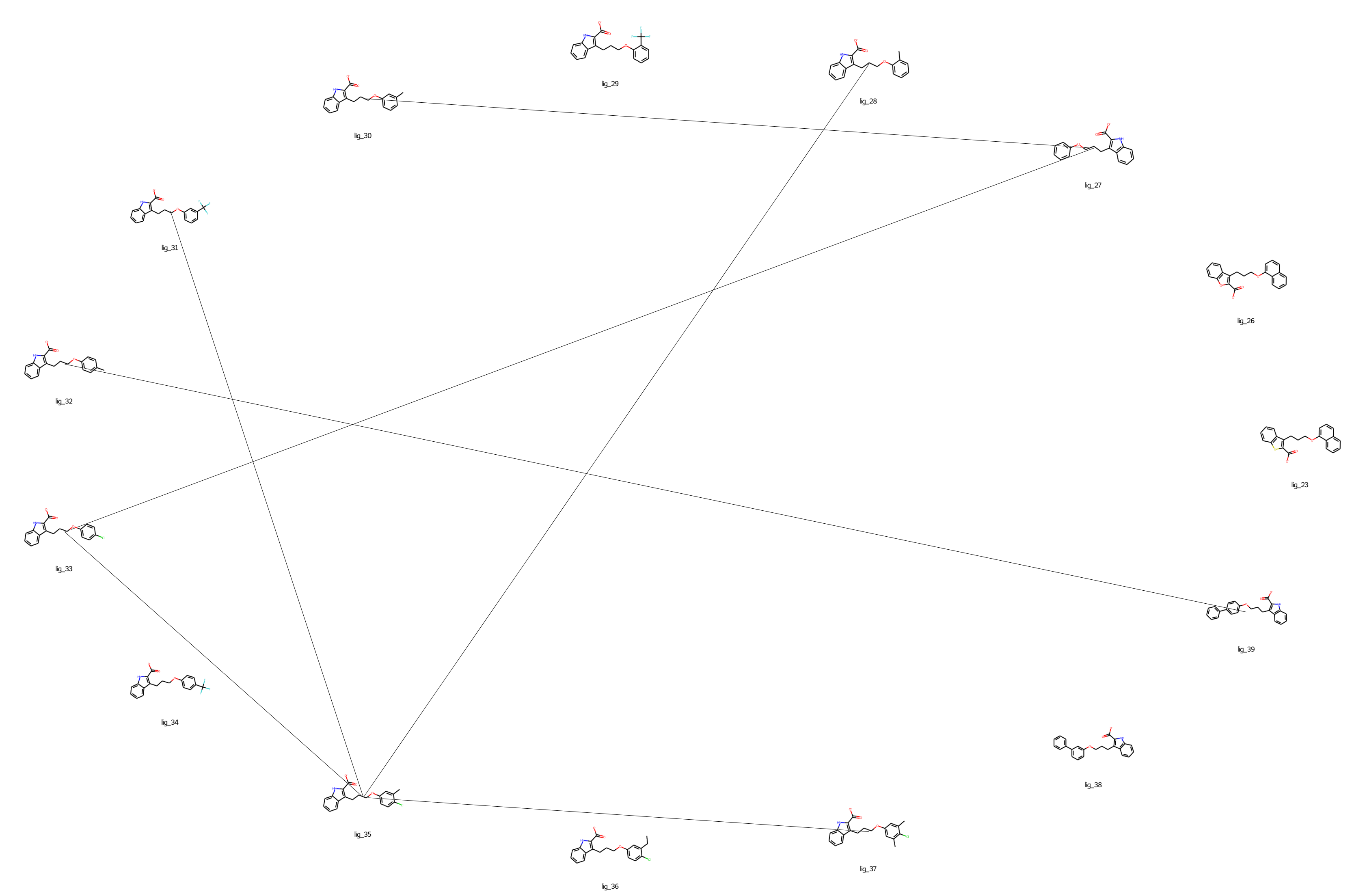
Finally, the set out of ligands and edges can be visualized in a graph:
[8]:
graph = mcl1.get_graph()
The LigandSet and Ligand class
The LigandSet consists of a Dict of Ligands which are availabe for one target. It is accessible via Target.get_ligand_set(), but can also be initialized directly.
[9]:
from plbenchmark import ligands
[10]:
mcl1_ligands = ligands.LigandSet('mcl1_sample')
HTML(mcl1_ligands.get_html())
/home/dhahn3/miniconda3/envs/plbenchmark/lib/python3.9/site-packages/pandas/core/dtypes/cast.py:1638: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
result[:] = values
[10]:
| name | smiles | measurement | DerivedMeasurement | ROMol | measurement | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| comment | error | type | unit | value | type | value | error | unit | Reference | ||||
| 0 | lig_23 | [H]c1c(c(c2c(c1[H])c(c(c(c2OC([H])([H])C([H])([H])C([H])([H])C3=C(Sc4c3c(c(c(c4[H])[H])[H])[H])C(=O)[O-])[H])[H])[H])[H])[H] | Table 2, entry 23 | 30 nanomolar | ki | nanomolar | 370 nanomolar | dg | -8.83 kilocalorie / mole | 0.05 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 1 | lig_26 | [H]c1c(c(c2c(c1[H])c(c(c(c2OC([H])([H])C([H])([H])C([H])([H])C3=C(Oc4c3c(c(c(c4[H])[H])[H])[H])C(=O)[O-])[H])[H])[H])[H])[H] | Table 2, entry 26 | 0.44 micromolar | ki | micromolar | 1.0 micromolar | dg | -8.24 kilocalorie / mole | 0.26 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 2 | lig_27 | [H]c1c(c(c(c(c1[H])[H])OC([H])([H])C([H])([H])C([H])([H])C2=C(N(c3c2c(c(c(c3[H])[H])[H])[H])[H])C(=O)[O-])[H])[H] | Table 3, entry 27 | 0.0071 millimolar | ki | millimolar | 0.035 millimolar | dg | -6.12 kilocalorie / mole | 0.12 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 3 | lig_28 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3C([H])([H])[H])[H])[H])[H])[H])[H])[H] | Table 3, entry 28, manually converted | 0.03 kilocalorie / mole | dg | kilocalorie / mole | -6.62 kilocalorie / mole | dg | -6.62 kilocalorie / mole | 0.03 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 4 | lig_29 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3C(F)(F)F)[H])[H])[H])[H])[H])[H] | Table 3, entry 29, manually converted | 120.0 calorie / mole | dg | calories / mole | -6940.0 calorie / mole | dg | -6.94 kilocalorie / mole | 0.12 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 5 | lig_30 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])[H])[H])[H])[H])[H] | Table 3, entry 30, manually converted | 0.6 micromolar | ic50 | micromolar | 1.9 micromolar | dg | -7.85 kilocalorie / mole | 0.19 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 6 | lig_31 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C(F)(F)F)[H])[H])[H])[H])[H] | Table 3, entry 31, manually converted | 80 nanomolar | ic50 | nanomolar | 1700 nanomolar | dg | -7.92 kilocalorie / mole | 0.03 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 7 | lig_32 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])[H])C([H])([H])[H])[H])[H])[H])[H] | Table 3, entry 32, manually converted | 0.08 dimensionless | pic50 | dimensionless | 4.8 dimensionless | dg | -6.59 kilocalorie / mole | 0.11 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 8 | lig_33 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])[H])Cl)[H])[H])[H])[H] | Table 3, entry 33, manually converted | 0.75 kilojoule / mole | dg | kilojoules / mole | -28.79 kilojoule / mole | dg | -6.880975143403441 kilocalorie / mole | 0.17925430210325047 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 9 | lig_34 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])[H])C(F)(F)F)[H])[H])[H])[H] | Table 3, entry 34 | 3.2 micromolar | ki | micromolar | 9.9 micromolar | dg | -6.87 kilocalorie / mole | 0.19 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 10 | lig_35 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)[H])[H])[H])[H] | Table 3, entry 35 | 0.14 micromolar | ki | micromolar | 0.38 micromolar | dg | -8.81 kilocalorie / mole | 0.22 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 11 | lig_36 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])C([H])([H])[H])Cl)[H])[H])[H])[H] | Table 3, entry 36 | 0.1 micromolar | ki | micromolar | 1.1 micromolar | dg | -8.18 kilocalorie / mole | 0.05 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 12 | lig_37 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)C([H])([H])[H])[H])[H])[H] | Table 3, entry 37 | 0.15 micromolar | ki | micromolar | 0.3 micromolar | dg | -8.95 kilocalorie / mole | 0.3 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 13 | lig_38 | [H]c1c(c(c(c(c1[H])[H])c2c(c(c(c(c2[H])OC([H])([H])C([H])([H])C([H])([H])C3=C(N(c4c3c(c(c(c4[H])[H])[H])[H])[H])C(=O)[O-])[H])[H])[H])[H])[H] | Table 3, entry 38 | 2.1 micromolar | ki | micromolar | 7.7 micromolar | dg | -7.02 kilocalorie / mole | 0.16 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
| 14 | lig_39 | [H]c1c(c(c(c(c1[H])[H])c2c(c(c(c(c2[H])[H])OC([H])([H])C([H])([H])C([H])([H])C3=C(N(c4c3c(c(c(c4[H])[H])[H])[H])[H])C(=O)[O-])[H])[H])[H])[H] | Table 3, entry 39 | 0.7 micromolar | ki | micromolar | 7.6 micromolar | dg | -7.03 kilocalorie / mole | 0.05 kilocalorie / mole | None | Friberg et al., J. Med. Chem. 2013 | |
The Ligand classes can be accessed from the LigandSet by their name. Each Ligand has information about experimental data, references, SMILES string and SDF file path of the docked structure. Additionally, there are functions to derive and process the primary data, which is then added to the pandas.Series as a new entry.
[11]:
lig_30 = mcl1_ligands['lig_30']
lig_27 = mcl1_ligands.get_ligand('lig_27')
The EdgeSet and Edge class
The EdgeSet contains a dict of Edges which are availabe for one target. It is accessible via Target.get_edge_set(), but can also be initialized directly.
[12]:
from plbenchmark import edges
[13]:
mcl1_edges = edges.EdgeSet('mcl1_sample')
HTML(mcl1_edges.get_html())
/home/dhahn3/miniconda3/envs/plbenchmark/lib/python3.9/site-packages/pandas/core/dtypes/cast.py:1638: UnitStrippedWarning: The unit of the quantity is stripped when downcasting to ndarray.
result[:] = values
[13]:
| ligand_a | ligand_b | name | Mol1 | Smiles1 | Mol2 | Smiles2 | exp. DeltaG [kcal/mol] | exp. Error [kcal/mol] | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | lig_28 | lig_35 | edge_28_35 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3C([H])([H])[H])[H])[H])[H])[H])[H])[H] | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)[H])[H])[H])[H] | -2.19 kilocalorie / mole | 0.22 kilocalorie / mole | ||
| 1 | lig_30 | lig_27 | edge_30_27 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])[H])[H])[H])[H])[H] | [H]c1c(c(c(c(c1[H])[H])OC([H])([H])C([H])([H])C([H])([H])C2=C(N(c3c2c(c(c(c3[H])[H])[H])[H])[H])C(=O)[O-])[H])[H] | 1.73 kilocalorie / mole | 0.22 kilocalorie / mole | ||
| 2 | lig_31 | lig_35 | edge_31_35 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C(F)(F)F)[H])[H])[H])[H])[H] | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)[H])[H])[H])[H] | -0.89 kilocalorie / mole | 0.22 kilocalorie / mole | ||
| 3 | lig_33 | lig_27 | edge_33_27 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])[H])Cl)[H])[H])[H])[H] | [H]c1c(c(c(c(c1[H])[H])OC([H])([H])C([H])([H])C([H])([H])C2=C(N(c3c2c(c(c(c3[H])[H])[H])[H])[H])C(=O)[O-])[H])[H] | 0.76 kilocalorie / mole | 0.22 kilocalorie / mole | ||
| 4 | lig_35 | lig_33 | edge_35_33 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)[H])[H])[H])[H] | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])[H])Cl)[H])[H])[H])[H] | 1.93 kilocalorie / mole | 0.28 kilocalorie / mole | ||
| 5 | lig_35 | lig_37 | edge_35_37 | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)[H])[H])[H])[H] | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])Cl)C([H])([H])[H])[H])[H])[H] | -0.14 kilocalorie / mole | 0.37 kilocalorie / mole | ||
| 6 | lig_39 | lig_32 | edge_39_32 | [H]c1c(c(c(c(c1[H])[H])c2c(c(c(c(c2[H])[H])OC([H])([H])C([H])([H])C([H])([H])C3=C(N(c4c3c(c(c(c4[H])[H])[H])[H])[H])C(=O)[O-])[H])[H])[H])[H] | [H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])[H])C([H])([H])[H])[H])[H])[H])[H] | 0.44 kilocalorie / mole | 0.12 kilocalorie / mole |
[14]:
mcl1_edges.keys()
[14]:
dict_keys(['edge_28_35', 'edge_30_27', 'edge_31_35', 'edge_33_27', 'edge_35_33', 'edge_35_37', 'edge_39_32'])
The Edge classes can be accessed from the EdgeSet by their name. They are lightweight and provide only access to a pandas.DataFrame and a Dict:
[15]:
edge_30_27 = mcl1_edges.get_edge('edge_30_27')
df = edge_30_27.get_dataframe()
edge_30_27.get_dict()
[15]:
{'ligand_a': 'lig_30',
'ligand_b': 'lig_27',
'name': 'edge_30_27',
'Mol1': <rdkit.Chem.rdchem.Mol at 0x7f1a3046e8e0>,
'Smiles1': '[H]c1c(c(c2c(c1[H])C(=C(N2[H])C(=O)[O-])C([H])([H])C([H])([H])C([H])([H])Oc3c(c(c(c(c3[H])C([H])([H])[H])[H])[H])[H])[H])[H]',
'Mol2': <rdkit.Chem.rdchem.Mol at 0x7f1a30460700>,
'Smiles2': '[H]c1c(c(c(c(c1[H])[H])OC([H])([H])C([H])([H])C([H])([H])C2=C(N(c3c2c(c(c(c3[H])[H])[H])[H])[H])C(=O)[O-])[H])[H]',
'exp. DeltaG [kcal/mol]': 1.73 <Unit('kilocalorie / mole')>,
'exp. Error [kcal/mol]': 0.22 <Unit('kilocalorie / mole')>}
[ ]: