Getting started with PLBenchmarks¶
from PLBenchmarks import targets
from IPython.core.display import HTML
Get the whole set of targets in the dataset¶
# it is initialized from the `PLBenchmarks/data/targets.yml` file
tgtset = targets.targetSet()
# to see which targets are available, one can get a list of names
tgtset.getNames()
['jnk1',
'pde2',
'thrombin',
'p38',
'ptp1b',
'galectin',
'cdk2',
'cmet',
'mcl1']
The targetSet is a dict, but can be converted to a
pandas.DataFrame or a html string via
targetSet.getDF(columns=None) or
targetSet.getHTML(columns=None). The default None for
columns means that all columns are printed. One can also define a
subset of columns as a list:
HTML(tgtset.getHTML(columns=['name', 'fullname', 'pdb', 'references', 'numLigands', 'minDG', 'maxDG', 'associated_sets']))
| name | fullname | pdb | references | numLigands | minDG | maxDG | associated_sets | |
|---|---|---|---|---|---|---|---|---|
| 0 | jnk1 | c-Jun N-terminal kinase 1 | 2GMX | [{'measurement': None}, {'calculation': ['10.1021/ja512751q', 'acs.jcim.9b00105']}] | 21 | -10.78111762414039 kcal/mol | -7.353005882502171 kcal/mol | [Schrodinger JACS] |
| 1 | pde2 | phosphodiesterase 2 | 4D08,4D09,6EZF | [{'measurement': ['10.1021/ml500262u', '10.1021/ja404449g']}, {'calculation': ['10.1038/s41598-018-23039-5']}] | 21 | -12.01124485346444 kcal/mol | -8.812821938199054 kcal/mol | None |
| 2 | thrombin | thrombin | 2ZFF | [{'measurement': None}, {'calculation': ['10.1021/ja512751q', '10.1021/acs.jcim.9b00105']}] | 11 | -9.177820267686423 kcal/mol | -7.480879541108987 kcal/mol | [Schrodinger JACS] |
| 3 | p38 | p38 alpha MAP kinase | 1OUY, 3FLY | [{'measurement': '10.1021/jm101423y'}, {'calculation': '10.1021/ja512751q, 10.1021/acs.jcim.9b00105'}] | 34 | -12.354423277849138 kcal/mol | -8.546808630776018 kcal/mol | [Schrodinger JACS] |
| 4 | ptp1b | protein-tyrosine phosphatase 1B | 2QBS | [{'measurement': None}, {'calculation': '10.1021/ja512751q, 10.1021/acs.jcim.9b00105'}] | 23 | -12.584340587592953 kcal/mol | -7.409826188396777 kcal/mol | [Schrodinger JACS] |
| 5 | galectin | galectin-3C | 5E89,5E8A,5E88 | [{'measurement': '10.1002/cbic.201600285'}, {'calculation': '10.1007/s10822-018-0110-5'}] | 8 | 0.0 kcal/mol | 0.0 kcal/mol | None |
| 6 | cdk2 | cyclin-dependent kinase 2 | 1H1Q, 2WEV | [{'measurement': '10.1021/ci5004027'}, {'calculation': '10.1021/ja512751q, 10.1021/acs.jcim.9b00105'}] | 16 | -11.349056331748212 kcal/mol | -7.09348579743778 kcal/mol | [Schrodinger JACS] |
| 7 | cmet | tyrosine-protein kinase Met; hepatocyte growth factor receptor (HGFR) | 4R1Y | [{'measurement': '10.1016/j.bmcl.2015.02.002'}, {'calculation': None}] | 12 | 0.0 kcal/mol | 0.0 kcal/mol | [Merck KGaA FEP Benchmarks, YANK Benchmarks] |
| 8 | mcl1 | Induced myeloid leukemia cell differentiation protein Mcl-1 | 4HW3 | [{'measurement': None}, {'calculation': '10.1021/ja512751q, 10.1021/acs.jcim.9b00105'}] | 42 | 0.0 kcal/mol | 0.0 kcal/mol | [Schrodinger JACS] |
A target can be accessed with its name in two ways
jnk1 = tgtset['jnk1']
pde2 = tgtset.getTarget('pde2')
The target class¶
contains all the available information about one target of PLBenchmarks.
It also has two member variables, _ligandSet and _edgeSet, which
contain the information about the available ligand and edges of the
respective target. A target can either be accessed from the
targetSet (see cell before) or initialized using its name via
jnk1 = targets.target('jnk1')
# The data in the column is stored in a pandas.Series and can be accessed via
jnk1.getDF(columns=None)
id 1
name jnk1
fullname c-Jun N-terminal kinase 1
netcharge xx
pdb 2GMX
ligands [lig_17124-1, lig_18624-1, lig_18625-1, lig_18...
references [{'measurement': None}, {'calculation': ['10.1...
comments None
associated_sets [Schrodinger JACS]
dtype: object
Access to the edgeSet and ligandSet in different formats is
achieved by
jnk1_ligands = jnk1.getLigandSet()
jnk1_ligands_df = jnk1.getLigandSetDF()
HTML(jnk1.getLigandSetHTML(columns = ['name', 'ROMol', 'measurement', 'DerivedMeasurement']))
| name | measurement | DerivedMeasurement | measurement | ROMol | |||||
|---|---|---|---|---|---|---|---|---|---|
| ic50 | doi | comment | e_ic50 | dg | e_dg | doi_html | |||
| 0 | lig_17124-1 | 77 nM | 10.1021/jm060199b | table 1 cmpd 6t | 38 nM | -9.76481161912498 kcal/mol | -10.185832695626143 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 1 | lig_18624-1 | 570 nM | 10.1021/jm060199b | table 1 cmpd 6e | 140 nM | -8.571396114764765 kcal/mol | -9.408403926601597 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 2 | lig_18625-1 | 1100 nM | 10.1021/jm060199b | table 1 cmpd 6f | 300 nM | -8.179461879338154 kcal/mol | -8.954045001030975 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 3 | lig_18626-1 | 300 nM | 10.1021/jm060199b | table 1 cmpd 6g | 70 nM | -8.954045001030975 kcal/mol | -9.821631925019588 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 4 | lig_18627-1 | 580 nM | 10.1021/jm060199b | table 1 cmpd 6h | 170 nM | -8.56102781892315 kcal/mol | -9.292655491812555 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 5 | lig_18628-1 | 400 nM | 10.1021/jm060199b | table 1 cmpd 6i | 120 nM | -8.782539885935572 kcal/mol | -9.500302701733787 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 6 | lig_18629-1 | 420 nM | 10.1021/jm060199b | table 1 cmpd 6j | 56 nM | -8.753453044861022 kcal/mol | -9.954661627304407 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 7 | lig_18630-1 | 190 nM | 10.1021/jm060199b | table 1 cmpd 6k | 4 nM | -9.22634699650534 kcal/mol | -11.527967281013156 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 8 | lig_18631-1 | 120 nM | 10.1021/jm060199b | table 1 cmpd 6l | 47 nM | -9.500302701733787 kcal/mol | -10.059111644635502 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 9 | lig_18632-1 | 210 nM | 10.1021/jm060199b | table 1 cmpd 6m | 95 nM | -9.166681043279013 kcal/mol | -9.639574994923331 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 10 | lig_18633-1 | 180 nM | 10.1021/jm060199b | table 1 cmpd 6n | 59 nM | -9.258579818411201 kcal/mol | -9.92355046515384 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 11 | lig_18634-1 | 45 nM | 10.1021/jm060199b | table 1 cmpd 6o | 3 nM | -10.085035815247183 kcal/mol | -11.699472396108563 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 12 | lig_18635-1 | 4400 nM | 10.1021/jm060199b | table 1 cmpd 6p | 670 nM | -7.353005882502171 kcal/mol | -8.475031685912471 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 13 | lig_18636-1 | 3000 nM | 10.1021/jm060199b | table 1 cmpd 6q | 1600 nM | -7.581331303492182 kcal/mol | -7.956083889099589 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 14 | lig_18637-1 | 35 nM | 10.1021/jm060199b | table 1 cmpd 6r | 16 nM | -10.234859923437579 kcal/mol | -10.701511284177176 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 15 | lig_18638-1 | 38 nM | 10.1021/jm060199b | table 1 cmpd 6s | 5 nM | -10.185832695626143 kcal/mol | -11.394937578728335 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 16 | lig_18639-1 | 69 nM | 10.1021/jm060199b | table 1 cmpd 6u | 14 nM | -9.83020994328735 kcal/mol | -10.78111762414039 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 17 | lig_18652-1 | 14 nM | 10.1021/jm060199b | table 3 cmpd 18b | 2 nM | -10.78111762414039 kcal/mol | -11.941195279431149 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 18 | lig_18658-1 | 74 nM | 10.1021/jm060199b | table 3 cmpd 20a | 11 nM | -9.788503292300433 kcal/mol | -10.924889274415738 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 19 | lig_18659-1 | 110 nM | 10.1021/jm060199b | table 3 cmpd 20b | 13 nM | -9.552175576876945 kcal/mol | -10.82529797985526 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
| 20 | lig_18660-1 | 400 nM | 10.1021/jm060199b | table 3 cmpd 20c | 45 nM | -8.782539885935572 kcal/mol | -10.085035815247183 kcal/mol | Szczepankiewicz et al., J. Med. Chem. 2006 | |
jnk1_edges = jnk1.getEdgeSet()
jnk1_edges_df = jnk1.getEdgeSetDF()
HTML(jnk1.getEdgeSetHTML())
| 0 | 1 | Mol1 | Mol2 | exp. DeltaG [kcal/mol] | |
|---|---|---|---|---|---|
| 0 | 17124-1 | 18634-1 | -0.32 | ||
| 1 | 18626-1 | 18624-1 | 0.38 | ||
| 2 | 18636-1 | 18625-1 | -0.60 | ||
| 3 | 18632-1 | 18624-1 | 0.60 | ||
| 4 | 18635-1 | 18625-1 | -0.83 | ||
| 5 | 18626-1 | 18658-1 | -0.83 | ||
| 6 | 18639-1 | 18658-1 | 0.04 | ||
| 7 | 18626-1 | 18625-1 | 0.77 | ||
| 8 | 18638-1 | 18658-1 | 0.40 | ||
| 9 | 18628-1 | 18624-1 | 0.21 | ||
| 10 | 18631-1 | 18660-1 | 0.72 | ||
| 11 | 18638-1 | 18634-1 | 0.10 | ||
| 12 | 18626-1 | 18632-1 | -0.21 | ||
| 13 | 18626-1 | 18630-1 | -0.27 | ||
| 14 | 18631-1 | 18624-1 | 0.93 | ||
| 15 | 18629-1 | 18627-1 | 0.19 | ||
| 16 | 18634-1 | 18637-1 | -0.15 | ||
| 17 | 18626-1 | 18627-1 | 0.39 | ||
| 18 | 18631-1 | 18652-1 | -1.28 | ||
| 19 | 18637-1 | 18631-1 | 0.73 | ||
| 20 | 18626-1 | 18634-1 | -1.13 | ||
| 21 | 18633-1 | 18624-1 | 0.69 | ||
| 22 | 17124-1 | 18631-1 | 0.26 | ||
| 23 | 18627-1 | 18630-1 | -0.67 | ||
| 24 | 18659-1 | 18634-1 | -0.53 | ||
| 25 | 18636-1 | 18624-1 | -0.99 | ||
| 26 | 18626-1 | 18628-1 | 0.17 | ||
| 27 | 18626-1 | 18660-1 | 0.17 | ||
| 28 | 18626-1 | 18659-1 | -0.60 | ||
| 29 | 18639-1 | 18634-1 | -0.25 | ||
| 30 | 18635-1 | 18624-1 | -1.22 |
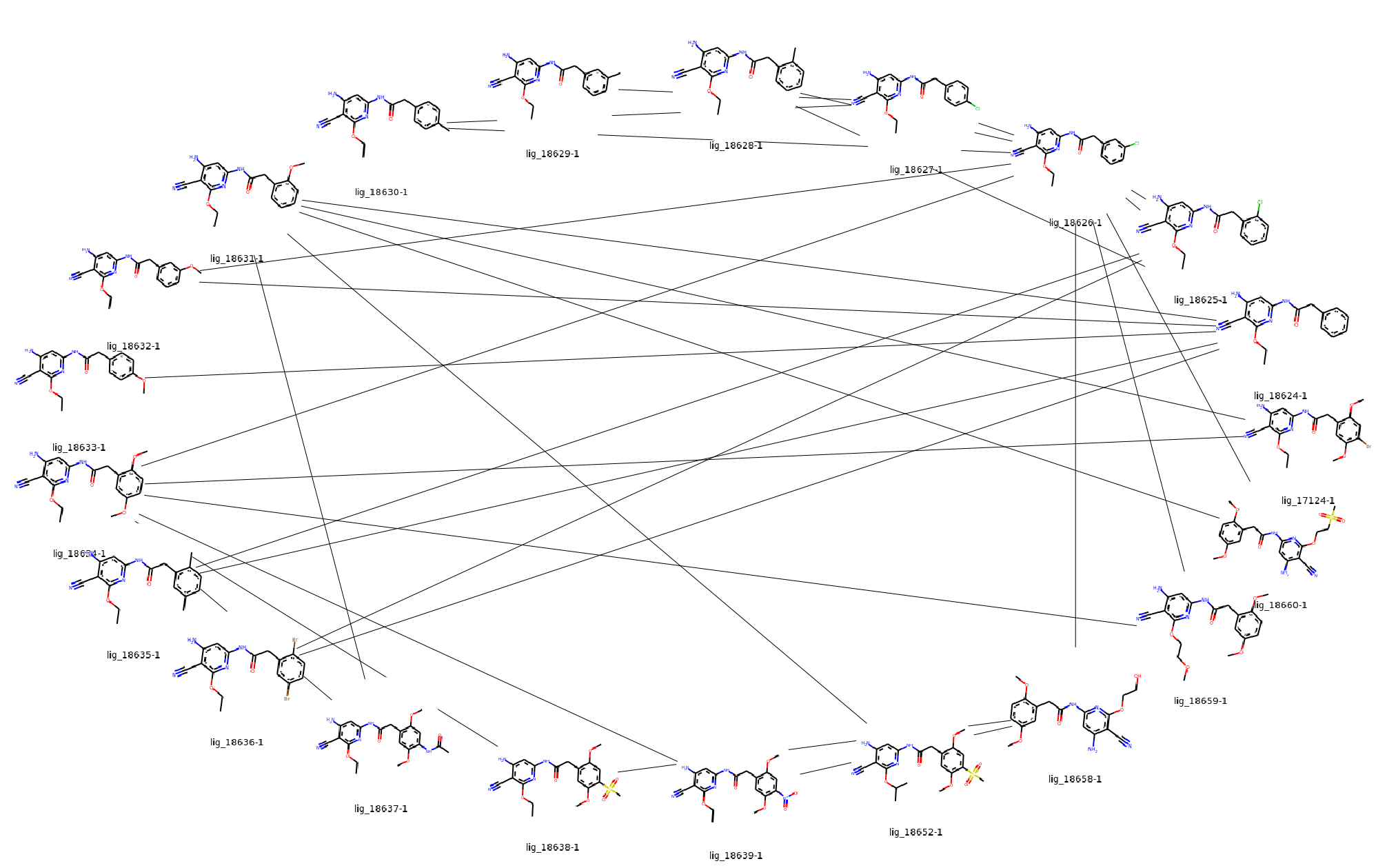
Finally, the set out of ligands and edges can be visualized in a graph:
graph = jnk1.getGraph()
/opt/anaconda3/envs/off-demo/lib/python3.7/site-packages/networkx/drawing/nx_pylab.py:579: MatplotlibDeprecationWarning:
The iterable function was deprecated in Matplotlib 3.1 and will be removed in 3.3. Use np.iterable instead.
if not cb.iterable(width):
The ligandSet and ligand class¶
The ligandSet consists of a dict of ligands which are
availabe for one target. It is accessible via target.getLigandSet(),
but can also be initialized directly.
from PLBenchmarks import ligands
thrombin_ligands = ligands.ligandSet('thrombin')
HTML(thrombin_ligands.getHTML())
| name | smiles | docked | measurement | DerivedMeasurement | measurement | ROMol | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dg | dh | tds | doi | comment | e_dg | e_dh | e_tds | dg | e_dg | doi_html | |||||
| 0 | lig_1a | c1ccc(cc1)C[C@H](C(=O)N2CCC[C@H]2C(=O)NCc3cccc(c3)F)[NH3+] | 03_docked/lig_1a/lig_1a.sdf | -31.3 kJ/mol | -13.1 kJ/mol | -18.2 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 1a | 0.2 kJ/mol | 0.9 kJ/mol | 0.7 kJ/mol | -7.480879541108987 kcal/mol | 0.04780114722753346 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
| 1 | lig_1b | c1ccc(cc1)C[C@H](C(=O)N2CCC[C@H]2C(=O)NCc3cccc(c3)Cl)[NH3+] | 03_docked/lig_1b/lig_1b.sdf | -35.4 kJ/mol | -37.1 kJ/mol | 1.7 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 1b | 0.8 kJ/mol | 1.1 kJ/mol | 0.3 kJ/mol | -8.460803059273422 kcal/mol | 0.19120458891013384 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
| 2 | lig_1c | c1ccc(cc1)C[C@H](C(=O)N2CCC[C@H]2C(=O)NCc3cccc(c3)Br)[NH3+] | 03_docked/lig_1c/lig_1c.sdf | -35.8 kJ/mol | -34.5 kJ/mol | -1.3 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 1c | 0.7 kJ/mol | 0.4 kJ/mol | 0.3 kJ/mol | -8.556405353728488 kcal/mol | 0.1673040152963671 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
| 3 | lig_1d | c1ccc(cc1)C[C@H](C(=O)N2CCC[C@H]2C(=O)NCc3cccc(c3)I)[NH3+] | 03_docked/lig_1d/lig_1d.sdf | -34.5 kJ/mol | -38.0 kJ/mol | -3.5 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 1d | 0.3 kJ/mol | 1.1 kJ/mol | 0.8 kJ/mol | -8.24569789674952 kcal/mol | 0.07170172084130018 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
| 4 | lig_3a | Cc1cccc(c1)CNC(=O)[C@@H]2CCCN2C(=O)[C@@H](Cc3ccccc3)[NH3+] | 03_docked/lig_3a/lig_3a.sdf | -34.8 kJ/mol | -28.5 kJ/mol | -6.3 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 3a | 0.6 kJ/mol | 0.7 kJ/mol | 1.1 kJ/mol | -8.317399617590821 kcal/mol | 0.14340344168260036 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
| 5 | lig_3b | CCc1cccc(c1)CNC(=O)[C@@H]2CCCN2C(=O)[C@@H](Cc3ccccc3)[NH3+] | 03_docked/lig_3b/lig_3b.sdf | -32.9 kJ/mol | -16.5 kJ/mol | -16.4 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 3b | 0.5 kJ/mol | 0.9 kJ/mol | 0.4 kJ/mol | -7.863288718929254 kcal/mol | 0.11950286806883365 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
| 6 | lig_5 | c1ccc(cc1)C[C@H](C(=O)N2CCC[C@H]2C(=O)NCc3ccccc3)[NH3+] | 03_docked/lig_5/lig_5.sdf | -31.7 kJ/mol | -13.6 kJ/mol | -18.1 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 5 | 0.0 kJ/mol | 0.0 kJ/mol | 0.0 kJ/mol | -7.576481835564053 kcal/mol | 0.0 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
| 7 | lig_6a | c1ccc(cc1)C[C@H](C(=O)N2CCC[C@H]2C(=O)NCc3cc(ccc3Cl)Cl)[NH3+] | 03_docked/lig_6a/lig_6a.sdf | -38.4 kJ/mol | -41.3 kJ/mol | 2.9 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 6a | 0.2 kJ/mol | 0.4 kJ/mol | 0.3 kJ/mol | -9.177820267686423 kcal/mol | 0.04780114722753346 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
| 8 | lig_6b | Cc1ccc(cc1CNC(=O)[C@@H]2CCCN2C(=O)[C@@H](Cc3ccccc3)[NH3+])Cl | 03_docked/lig_6b/lig_6b.sdf | -37.2 kJ/mol | -33.5 kJ/mol | -3.7 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 6b | 0.5 kJ/mol | 1.7 kJ/mol | 2.2 kJ/mol | -8.891013384321225 kcal/mol | 0.11950286806883365 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
| 9 | lig_6e | c1ccc(cc1)C[C@H](C(=O)N2CCC[C@H]2C(=O)NCc3cc(ccc3F)Cl)[NH3+] | 03_docked/lig_6e/lig_6e.sdf | -37.3 kJ/mol | -41.0 kJ/mol | 3.8 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 6e | 0.3 kJ/mol | 2.1 kJ/mol | 2.2 kJ/mol | -8.914913957934989 kcal/mol | 0.07170172084130018 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
| 10 | lig_7a | Cc1ccc(c(c1)CNC(=O)[C@@H]2CCCN2C(=O)[C@@H](Cc3ccccc3)[NH3+])C | 03_docked/lig_7a/lig_7a.sdf | -34.4 kJ/mol | -31.9 kJ/mol | -2.5 kJ/mol | 10.1016/j.jmb.2009.04.051 | Table 1 cmpd 7a | 0.1 kJ/mol | 1.3 kJ/mol | 1.3 kJ/mol | -8.221797323135755 kcal/mol | 0.02390057361376673 kcal/mol | Baum et al., Journal of Molecular Biology 2009 | |
The ligand classes can be accessed from the ligandSet by their
name. Each ligand has information about experimental data,
references, SMILES string and SDF file path of the docked structure.
Additionally, there are functions to derive and process the primary
data, which is then added to the pandas.Series as a new entry.
lig_6e = thrombin_ligands['lig_6e']
lig_1a = thrombin_ligands.getLigand('lig_6e')
The edgeSet and edge class¶
The edgeSet contains a dict of edges which are availabe
for one target. It is accessible via target.getEdgeSet(), but can
also be initialized directly.
from PLBenchmarks import edges
pde2_edges = edges.edgeSet('pde2')
HTML(pde2_edges.getHTML())
| 0 | 1 | Mol1 | Mol2 | exp. DeltaG [kcal/mol] | |
|---|---|---|---|---|---|
| 0 | 49220392 | 49137530 | 0.10 | ||
| 1 | 49932714 | 49137530 | -1.30 | ||
| 2 | 49582468 | 49137530 | -0.84 | ||
| 3 | 49396360 | 49137530 | -0.85 | ||
| 4 | 50181001 | 49137530 | -1.66 | ||
| 5 | 49585367 | 49137530 | -1.56 | ||
| 6 | 49220392 | 49175828 | -0.74 | ||
| 7 | 49220548 | 49220392 | -0.03 | ||
| 8 | 49220548 | 49932129 | 1.56 | ||
| 9 | 49582468 | 49932129 | 0.66 | ||
| 10 | 49396360 | 49175828 | -1.69 | ||
| 11 | 49175828 | 49580115 | 1.89 | ||
| 12 | 49220548 | 49137374 | -0.04 | ||
| 13 | 49220548 | 49580115 | 1.13 | ||
| 14 | 49396360 | 49220548 | -0.92 | ||
| 15 | 49932714 | 49582390 | -0.91 | ||
| 16 | 49396360 | 49582390 | -0.45 | ||
| 17 | 50181001 | 49582390 | -1.26 | ||
| 18 | 50107616 | 49582390 | -1.33 | ||
| 19 | 48168913 | 48271249 | -0.45 | ||
| 20 | 49072088 | 48271249 | -2.06 | ||
| 21 | 50107616 | 48271249 | -1.43 | ||
| 22 | 49137374 | 48271249 | 0.41 | ||
| 23 | 49932714 | 49175789 | -1.25 | ||
| 24 | 49932714 | 49580115 | -0.25 | ||
| 25 | 49932714 | 49582468 | -0.47 | ||
| 26 | 48168913 | 49585367 | 0.81 | ||
| 27 | 50107616 | 49585367 | -0.16 | ||
| 28 | 48168913 | 48022468 | -1.54 | ||
| 29 | 43249674 | 48022468 | -0.19 | ||
| 30 | 48009208 | 43249674 | -0.75 | ||
| 31 | 43249674 | 49175789 | 0.65 | ||
| 32 | 49175789 | 49072088 | 2.31 | ||
| 33 | 48009208 | 49137374 | -0.27 |
pde2_edges.keys()
dict_keys(['edge_49220392_49137530', 'edge_49932714_49137530', 'edge_49582468_49137530', 'edge_49396360_49137530', 'edge_50181001_49137530', 'edge_49585367_49137530', 'edge_49220392_49175828', 'edge_49220548_49220392', 'edge_49220548_49932129', 'edge_49582468_49932129', 'edge_49396360_49175828', 'edge_49175828_49580115', 'edge_49220548_49137374', 'edge_49220548_49580115', 'edge_49396360_49220548', 'edge_49932714_49582390', 'edge_49396360_49582390', 'edge_50181001_49582390', 'edge_50107616_49582390', 'edge_48168913_48271249', 'edge_49072088_48271249', 'edge_50107616_48271249', 'edge_49137374_48271249', 'edge_49932714_49175789', 'edge_49932714_49580115', 'edge_49932714_49582468', 'edge_48168913_49585367', 'edge_50107616_49585367', 'edge_48168913_48022468', 'edge_43249674_48022468', 'edge_48009208_43249674', 'edge_43249674_49175789', 'edge_49175789_49072088', 'edge_48009208_49137374'])
The edge classes can be accessed from the edgeSet by their name.
They are lightweight and provide only access to a pandas.DataFrame
and a dict:
edge_49220392_49137530 = pde2_edges.getEdge('edge_49220392_49137530')
df = edge_49220392_49137530.getDF()
edge_49220392_49137530.getDict()
{'edge_49220392_49137530': ['lig_49220392', 'lig_49137530']}